This is the blogpost for the paper ‘Self-supervised Learning for Medical Image Analysis Using Image Context Restoration’.
Written by Liang Chen, Paul Bentley, Kensaku Mori, Kazunari Misawa, Michitaka Fujiwara, Daniel Rueckert
Introduction
Deep Learning methods have achieved great success in computer vision. Especially, CNNs have recently demonstrated impressive results in medical image domains such as disease classification[1] and organ segmentation[2]. Good deep learning model usually requires a decent amount of labels, but in many cases, the amount of unlabelled data is substantially more than the labelled ones. Also, the pre-trained models from the natural images are not useful on medical images since the intensity distribution is different. Besides, labelling natural images are easy, and just simple human knowledge is enough. However, the annotation for medical images requires expert knowledge.
So, how should we learn representations without labels?
The answer is simple; get supervision from the data or image itself. It means that we can achieve this by framing a supervised learning task in a particular form to predict only a subset of information using the rest. This is known as self-supervised learning.
What is self-supervised learning?
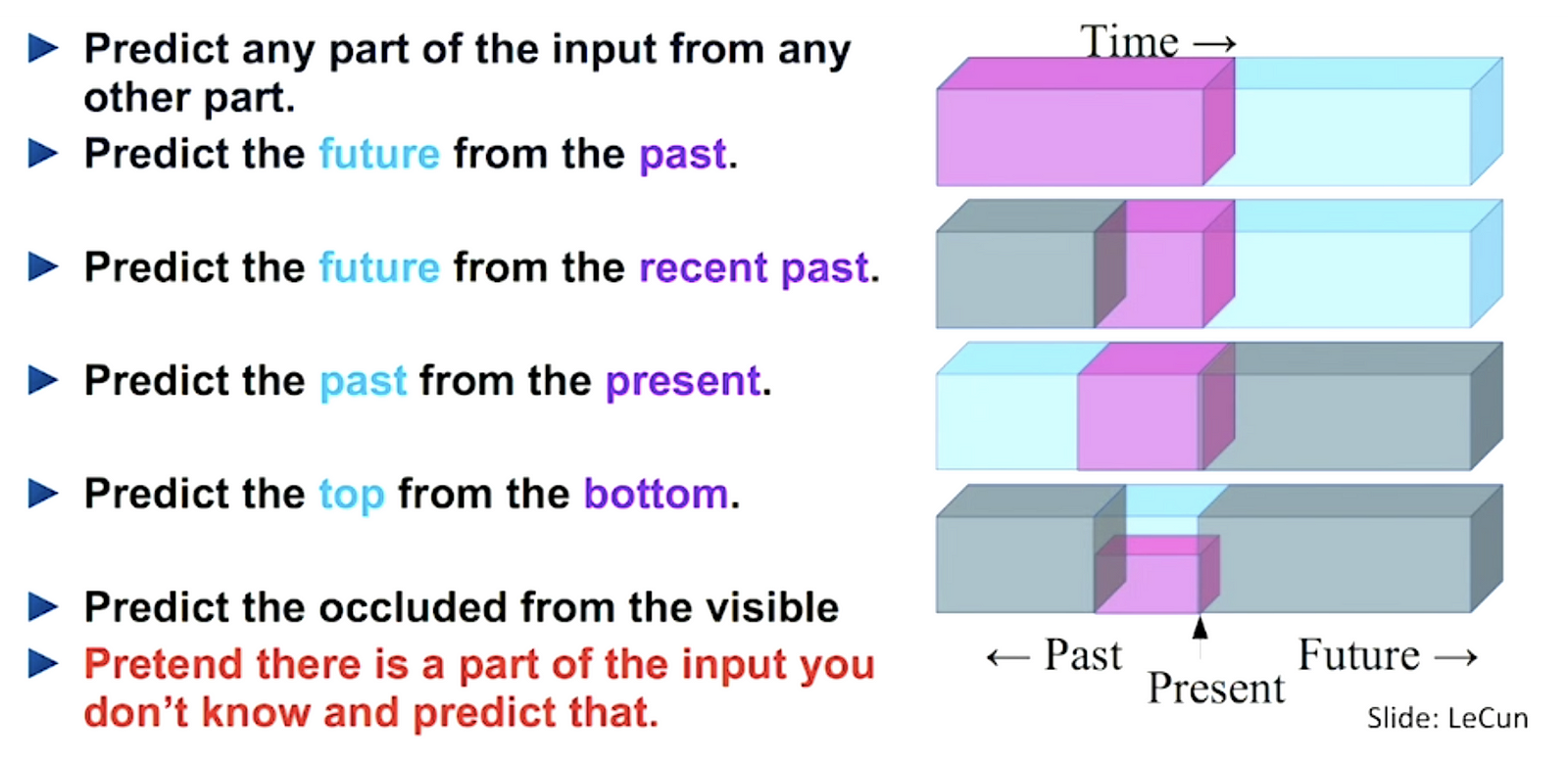
It is still supervised learning since it uses labelled data. It extracts and uses the naturally available relevant context. From that fact, a very large number of training instances with supervision are available. Thanks to this, pre-training a CNN based on such self-supervision results in useful weights to initialise the subsequent CNN based on data with limited manual labels. Fig. 1 shows us the summary of self-supervised learning.
Figure 1. A summary of self-supervised learning [3]
Since existing self-supervised learning strategies do not deliver a notable performance improvement on medical images, the authors propose another the self-supervision strategy which is called context restoration.
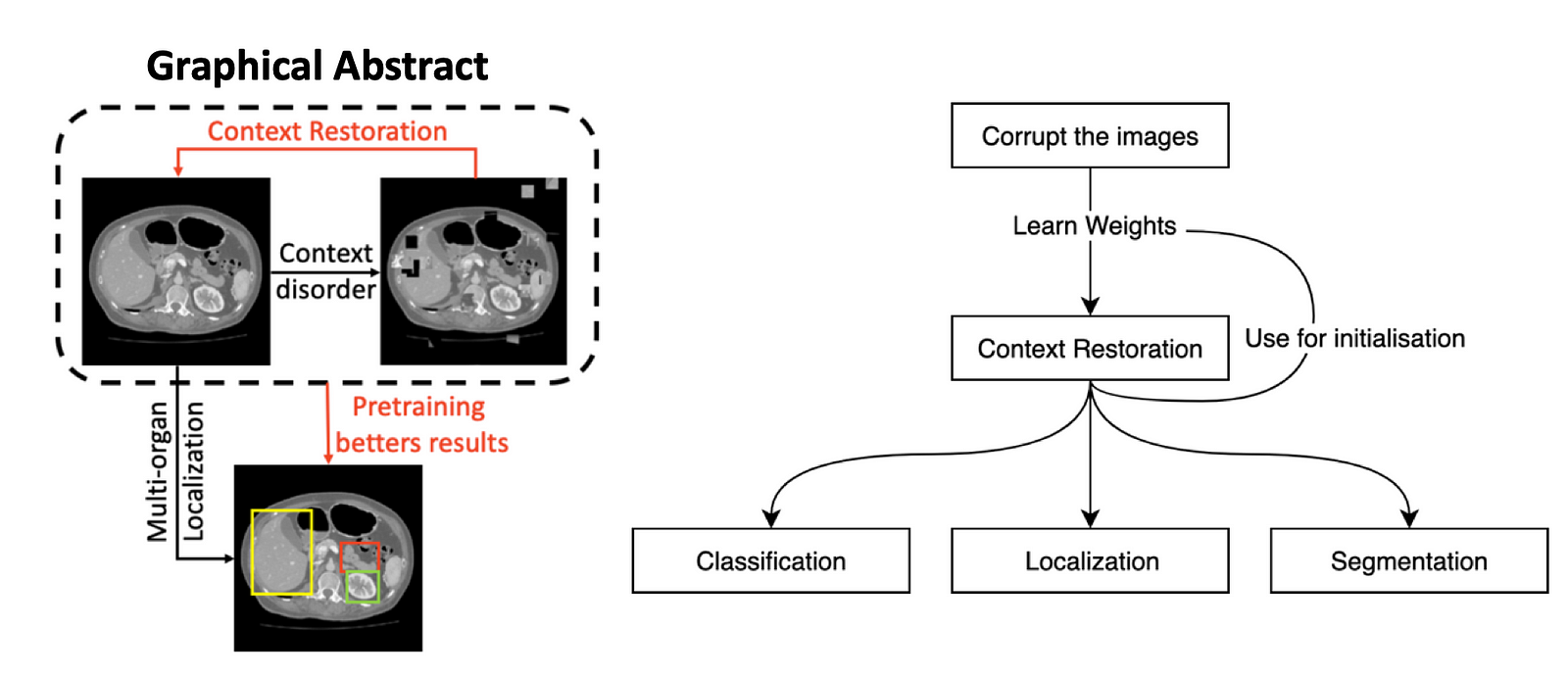
Figure 2. An overview of the context restoration method
Fig. 2 is a great overview of how the context restoration works. Basically, there are input images, then the algorithm corrupts them and tries to restore them. During this restoration process, it learns weights which will give us better results while working on subsequent tasks.
One of the self-supervised research, which is titled Unsupervised Visual Representation Learning by Context Prediction [4], predicts the relative position between a central patch and its surrounding patches in a 3 × 3 patch grid from the given image. For example, the right ear of a cat would be in the top-right position relative to the eyes of a cat. But it has three shortcomings: there can be multiple correct answers (e.g., cars or buildings), it still learning trivial features which are not useful in the medical images, and lastly, patches do not contain the information about the global context of images.
In terms of features learning, another self-supervised research proposed, which is named Context encoders: Feature learning by inpainting [5], is trained to fill in a missing piece in the image. Therefore, Reconstruct the original input while learning meaningful latent representation. Also, this one comes with some shortcomings: it changes the image intensity distribution. Thus, the resulting images belong to another domain, and the learned features may not be useful for images in the original domain.
Figure 3. Demonstration of the Relative Position(a) and Context Prediction(b) Methods [4, 5]
Methodology
The paper proposes a novel strategy for self-supervised which they term context restoration. The method is straightforward that select random two isolated small patches in given image and swap their context. Repeat these operation T times, which the intensity distribution is still preserved, but its spatial information is altered.
The paper validates the context restoration strategy in three common problems in medical imaging: classification, localisation, and segmentation. To scan plane detection in fetal 2D ultrasound images; to localise abdominal organs in CT images; to segment brain tumours in multi-modal MR images are used.
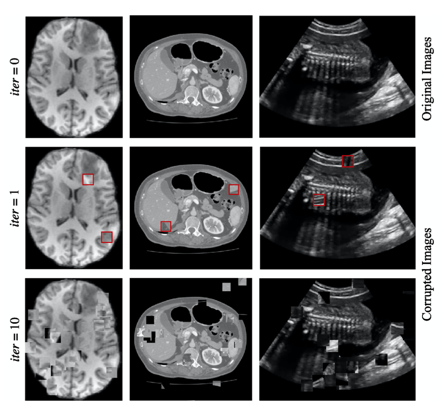
Figure 4. Generating training images for self-supervised context disordering: Brain T1 MR image, abdominal CT image, and 2D fetal ultrasound image, respectively. In figures in the second column, red boxes highlight the swapped patches after the first iteration.
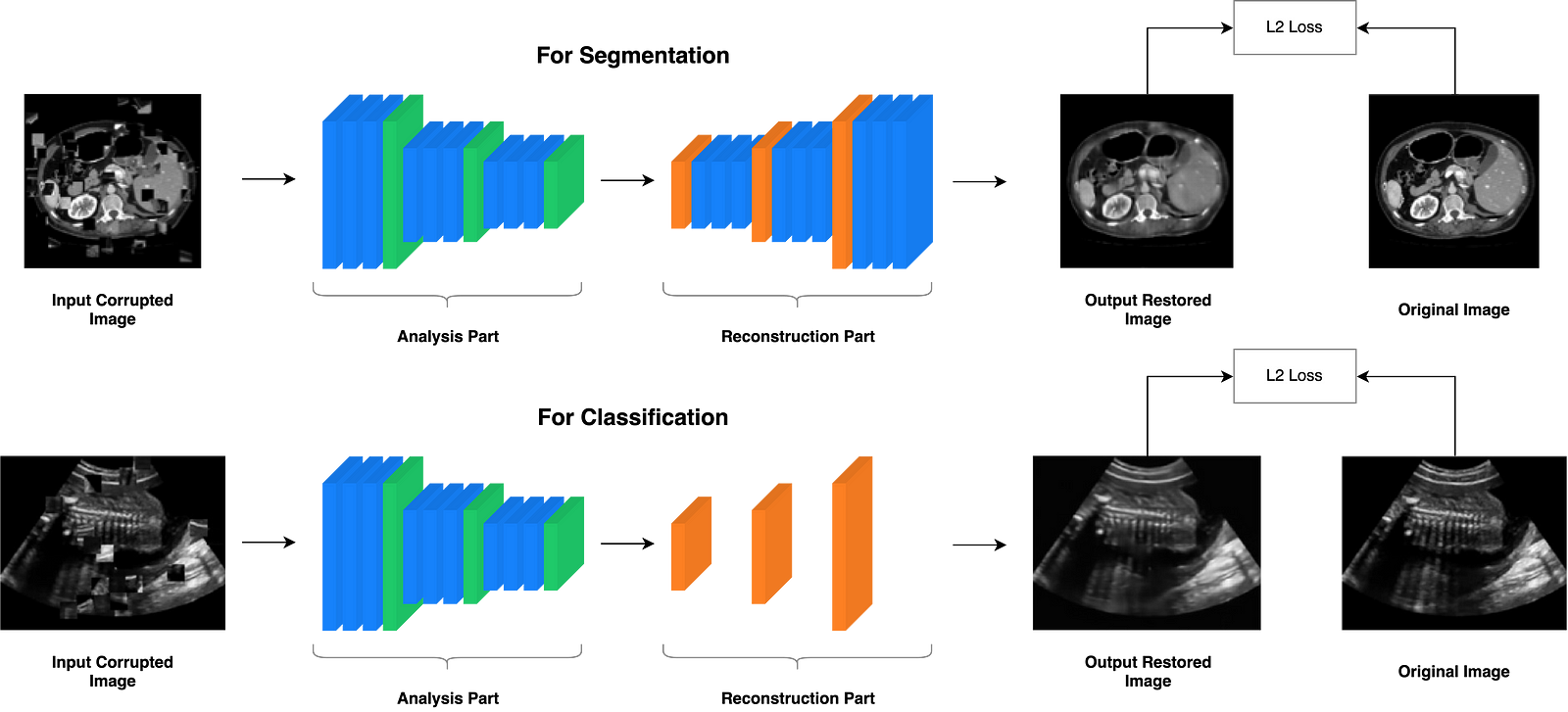
The proposed self-supervised learning strategy uses CNNs, and it consists of two parts: an analysis part and a reconstruction part. Fig. 3 shows an overview of the general architecture of feasible CNNs. The analysis part encodes given disordered images into feature maps, and the reconstruction part uses these feature maps to produce output images for correct context.
Analysis Part
This part includes stacks of convolutional units and downsampling units, which are extracting feature maps from given images.
Reconstruction Part
Here, it includes stacks of convolutional layers and upsampling layers, which restores the image. Reconstruction part is different for each subsequent task. For subsequent classification tasks, the simple structures such as a few deconvolution layers are preferred. For subsequent segmentation tasks, the complex structures consistent with the segmentation CNNs are preferred.
As a result, almost all the weights of the subsequent segmentation CNN can be initialised using those learned in the self-supervised pre-training.
Figure 5. General CNN architecture for the context restoration self-supervised learning.
Experimental Setup
The proposed self-supervision using context restoration task can be performed by CNNs on three different datasets, including brain MR images, abdominal CT images, and fetal US images, and they used the pre-trained CNNs for subsequent tasks such as classification, localisation, and segmentation, respectively. For each of these problems, a different CNN architecture and data is used:
Classification
The dataset consists of 2694 2D ultrasound examinations of fetuses (224×288) with gestational ages between 18 and 22 weeks [6].
The CNN for this classification problem is the SonoNet-64 which achieved the best performance in [6].
The performance of CNNs in this classification task is measured by the precision, recall, and the F1-score.
Localisation
A dataset of 3D abdominal CT image from 150 subjects is employed [7].
The dataset is randomly divided into two halves. The first half is used for training and validation, and the other half is used for testing.
The CNN for multi-organ localisation task is similar to the SonoNet [6]. It has one more stack of convolution and pooling layers than the SonoNet since the input images are 512×512, which is approximately twice larger than the processed 2D ultrasound frames in each side.
The performance is measured by computing the distances of the centroids and walls between bounding boxes.
Segmentation
The dataset of the BraTS 2017 challenge which consists of 285 subjects [8]. Each subject has MR images in multiple modalities, namely, native T1 (T1), post-contrast T1-weighted (T1-Gd), T2-weighted (T2), T2 fluid-attenuated inversion recovery (FLAIR).
142 out of the 285 images for training and validation and the remaining 143 ones used for testing.
The CNN used in this experiment is a 2D U-Net [9].
They use the same evaluation metrics in the BraTS 2017 challenge: Dice score, sensitivity, specificity, and Hausdorff distance.
They compared different self-supervised learning strategies, namely, random [6], random + augmentation, auto-encoder [10], self-supervision using patch relative position prediction [4], Jigsaw [11], and self-supervision using local context prediction [5] and the proposed context restoration.
Results and Discussion
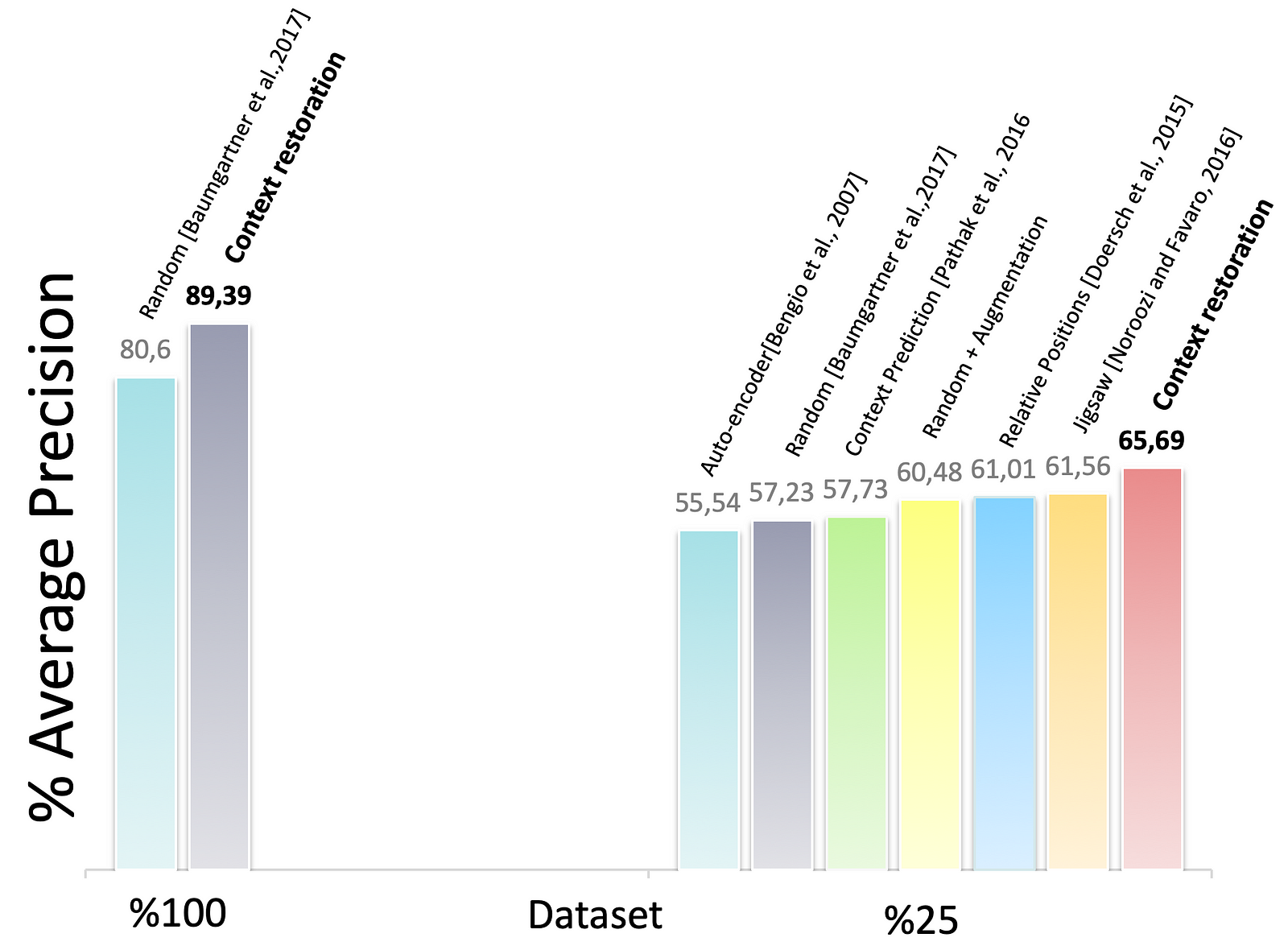
Figure 6. The classification of standard scan planes of fetal 2D ultrasound images
Fig. 6 demonstrates the results of the performance of the CNNs under different configurations. Thanks to self-supervised pretraining, the performance of CNNs when using small training datasets can be improved. The context restoration pretraining improves the SonoNet performance the most.
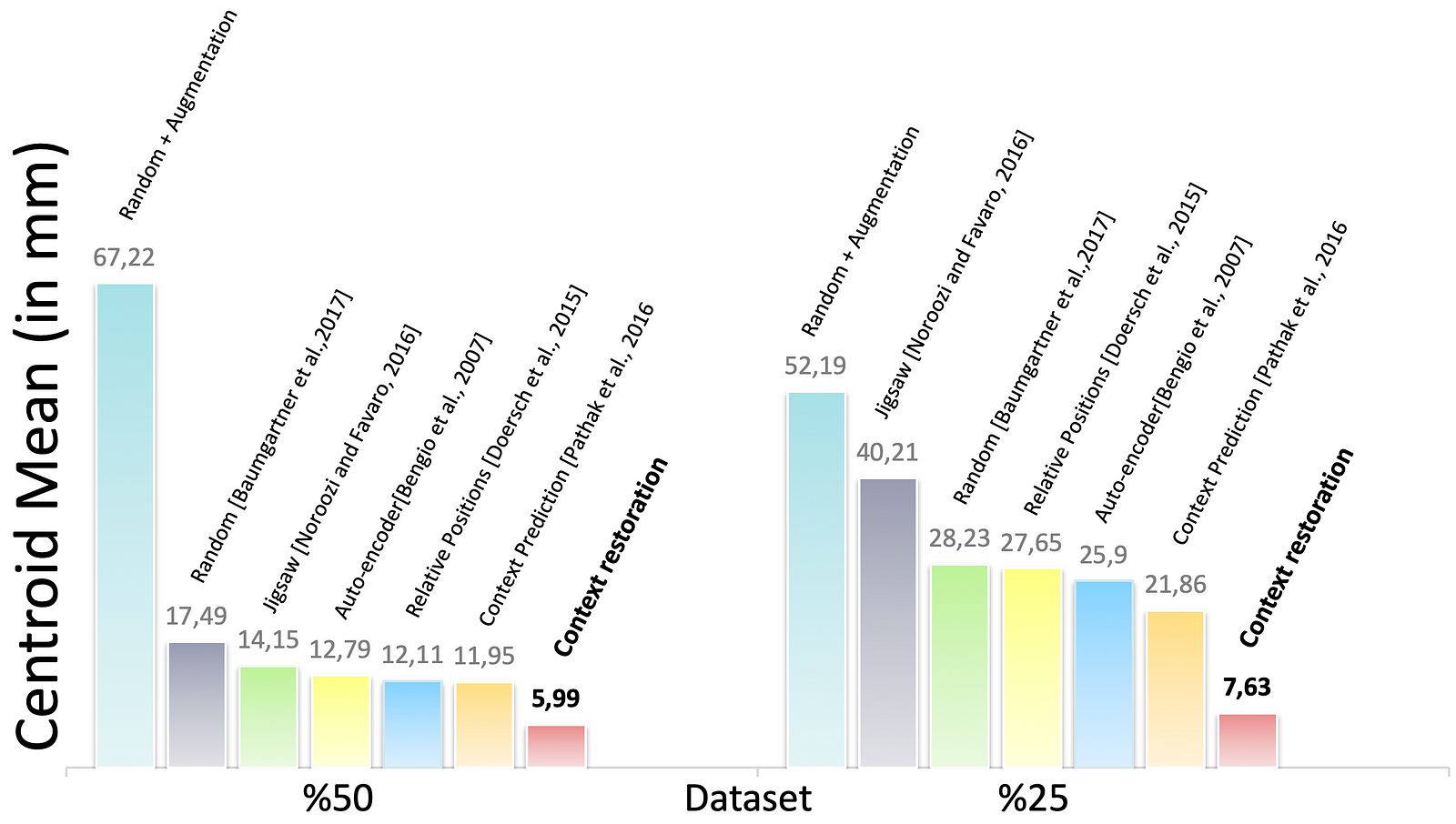
Figure 7. The performance of CNN solving the multi-organ localisation (Left Kidney in this result) problem in different training settings
Fig. 7 represents the localisation performance of the CNN in different training approaches. In some cases, the CNN using context restoration pre-training is comparable to or even better than none pre-training on more labelled training data. In terms of the left kidney, the CNN on half training data slightly outperforms that on all the training data. It is remarkable that if less training data leads to a significant decrease in results, self-supervised learning tends to improve the results significantly.
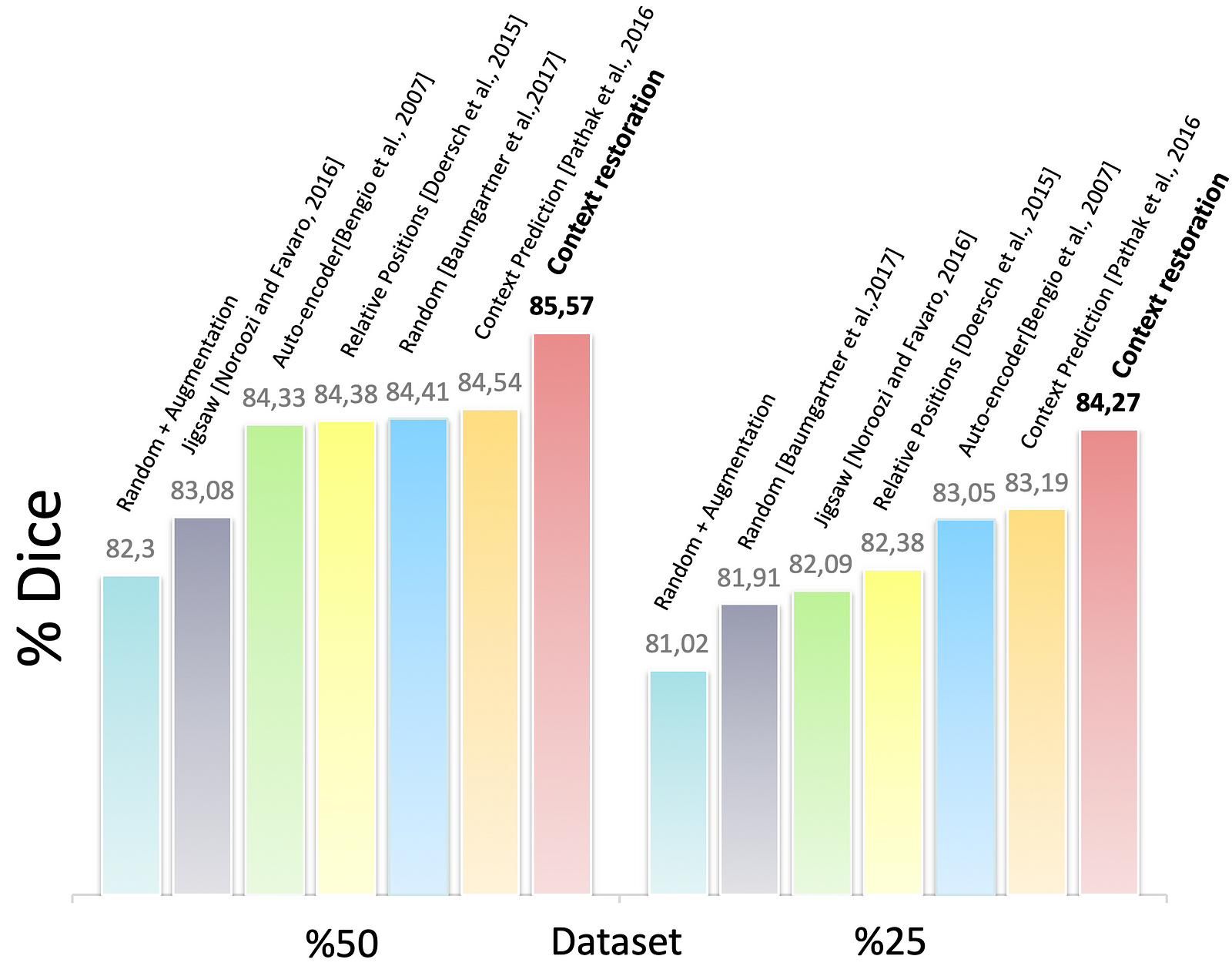
Figure 8. The segmentation results of the customised U-Nets in different training settings
Fig. 8 shows the results of the BraTS problem. The Dice score in the enhanced tumour core. And, it shows us to use half of the training dataset, the proposed self-supervision strategy results in a similar performance to using the whole training dataset. Again, we can see self-supervision based on context restoration offers a best pre-training approach for the segmentation task.
In this paper, the authors proposed a novel self-supervised learning strategy based on context restoration. This enables CNNs to learn useful image semantics without any labels and uses for the subsequent tasks. Therefore, we can conclude with the following key points:
- In all three tasks, which are classification, localisation, and segmentation, using context restoration pretraining performs better than others
- If reducing the training data causes significant performance decrease, the context restoration pretraining can improve the performance
- The context restoration has three notable features; learns semantic image features; these semantic features are useful for different subsequent tasks; implementation is simple and straightforward
- A shortcoming is that the L2 loss results in image blur.
Future Works
It is remarkable to explore more powerful self-supervised learning strategy so that the self-supervised pre-training can be as good as supervised pre-training in the future. In addition, the following questions arise:
- What would happen if a larger dataset is used? or optimised self-supervised learning model?
- How would affect the results if they could use 3x3 patches and swap randomly?
- Adding an additional model in order to restore faster?
- Using another loss function instead of L2, e.g., adversarial loss?
For further details on this work, check out the paper on ScienceDirect.
References
[1] Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., Summers, R. M., 2017. ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 3462–3471. URL https://arxiv.org/abs/1705.02315.
[2] Suk, H.-I., Lee, S.-W., Shen, D., Initiative, A. D. N., et al., 2014. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage 101, 569–582. URL https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4165842/.
[3] Yann LeCun, Self-supervised learning: could machines learn like humans, 2018. URL youtube.com.
[4] Doersch, C., Gupta, A., Efros, A. A., 2015. Unsupervised visual representation learning by context prediction. In: Proceedings of the IEEE International Conference on Computer Vision. pp. 1422–1430. URL https://arxiv.org/abs/1505.05192.
[5] Pathak, D., Krahenbuhl, P., Donahue, J., Darrell, T., Efros, A. A., 2016. Context encoders: Feature learning by inpainting. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 2536–2544. URL https://arxiv.org/abs/1604.07379.
[6] Baumgartner, C. F., Kamnitsas, K., Matthew, J., Fletcher, T. P., Smith, S., Koch, L. M., Kainz, B., Rueckert, D., 2017. SonoNet: real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Transactions on Medical Imaging 36 (11), 2204–2215. URL https://arxiv.org/abs/1612.05601.
[7] Tong, T., Wolz, R., Wang, Z., Gao, Q., Misawa, K., Fujiwara, M., Mori, K., Hajnal, J. V., Rueckert, D., 2015. Discriminative dictionary learning for abdominal multi-organ segmentation. Medical Image Analysis 23 (1), 92–104. URL https://www.ncbi.nlm.nih.gov/pubmed/25988490.
[8] Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R., et al., 2015. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Transactions on Medical Imaging 34 (10), 1993–2024. URL https://ieeexplore.ieee.org/document/6975210.
[9] Ronneberger, O., Fischer, P., Brox, T., 2015. U-net: convolutional networks for biomedical image segmentation. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 234–241. URL https://arxiv.org/abs/1505.04597.
[10] Bengio, Y., Lamblin, P., Popovici, D., Larochelle, H., 2007. Greedy layer-wise training of deep networks. In: Advances in Neural Information Processing Systems. pp. 153–160. URL https://papers.nips.cc/paper/3048-greedy-layer-wise-training-of-deep-networks.pdf.
[11] Noroozi, M., Favaro, P., 2016. Unsupervised learning of visual representations by solving jigsaw puzzles. In: Proceedings of the European Conference on Computer Vision. pp. 69–84. URL https://arxiv.org/abs/1603.09246.